Understanding cellular metabolism and tissue interactions
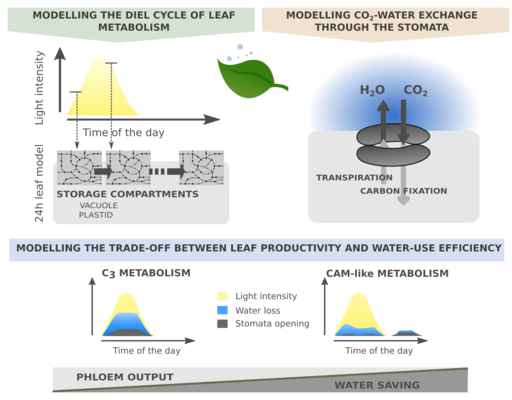
We aim to understand the constraints and capacities of leaf metabolism to develop crop plants that are better adapted to adverse future climates. Our focus is on photosynthetic diversity and the interplay between leaf metabolism and anatomy. We have previously used large-scale metabolic models to study CAM photosynthesis, alternative water-saving flux modes, and guard cell metabolism. Our current research explores the metabolic flux modes and cellular energetics of C3, C3-C4 intermediate, and C4 photosynthesis in the context of leaf anatomy. We are also interested in starch metabolism and its plasticity with respect to nitrogen availability and day lenghts.
¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯
Whole-plant metabolic models and plant life cycles

PlantEd — A serious game for education and outreach
We developed PlantEd, a strategy game for science communication and teaching. Players allocate biomass to grow leaves, branches, roots, and flowers, manage resources, and fend off herbivores to maximize seed biomass within the plant's life cycle. The game uses a whole-plant metabolic model and dynamic flux-balance analysis to simulate plant growth and survival. PlantEd is both educational and fun, teaching plant physiology basics and immersing players in the fascinating world of plants. This project is a collaboration with AG Szymanski (IPK Gatersleben, FZ Jülich).
On the scientific side, we use this framework to combine machine learning and metabolic modelling to study optimal resource allocation strategies in different environments and to develop multi-scale life cycle models to predict the ecological performance of Arabidopsis thaliana in different habitats and climates.

Integrating plant metabolic and architectural models
Plant architecture and metabolism mutually influence each other. To describe these dependencies, we have started to couple large-scale metabolic and functional-structural plant models. The latter provide a representation of whole-plant architecture and allow for the incorporation of meteorological datasets. We will employ our integrated model to better understand carbon and nitrogen partitioning and resource allocation towards different organs in grain crops under different and changing environmental scenarios.
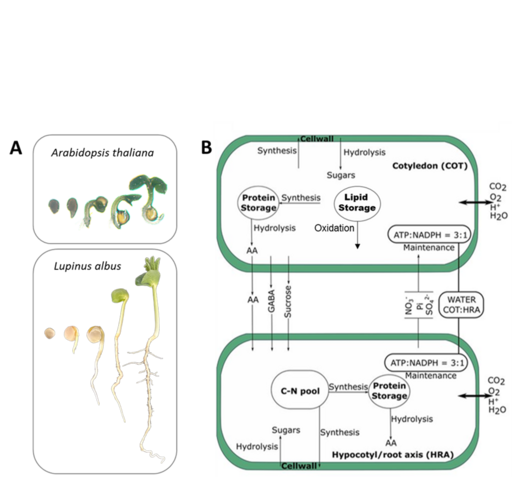
Flux modelling during seedling development and growth
During germination, plants depend on seed storage compounds for energy and macromolecule synthesis. Carbohydrates, lipids, and proteins act as major storage compounds. However, how storage compound usage is coordinated in various seed types remains poorly understood. In collaboration with AG Hildebrandt (UoC), we develop a time-resolved metabolic model to analyze carbon and nitrogen fluxes in the developing and growing seedling and parameterize this model with proteomics and metabolomics data from Arabidopsis thaliana and Lupinus albus seeds grown under ambient and adverse conditions. This study will shed light on metabolic adaptations for using lipids or proteins as major seed storage compounds.
¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯
Modelling plant-microbe interactions

In natural environments, plants coexist with a diverse array of microorganisms, including bacteria, fungi, oomycetes, archaea, and viruses. These microorganisms interact with each other and with the host plant in complex ways, which can be beneficial (mutualism, synergism, or commensalism), neutral, or even harmful (amensalism). Plants rely on their microbiome for essential traits, such as nutrient acquisition and tolerance to biotic and abiotic factors. These interactions are largely driven by metabolic exchanges or metabolite-derived signals and can thus be understood through models of metabolism and signaling. We have started to develop and apply computational methods to anaylse plant-microbe interactions and work closely with experimental partners. By elucidating these mechanisms, we will gain insights into what drives and shapes plant-microbial interactions, ultimately informing strategies for optimizing plant-microbe relationships.
¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯
Supporting the scientific community through open-source software

Modelling predictions strongly depend on the quality of the underlying metabolic network reconstruction. These network reconstructions comprise hundreds to thousands of reactions which are typically assembled from metabolic pathway databases and are thus sensitive to annotation errors and error propagation. To address this issue we are supplying open-source software packages which offer coherent and reproducible curation workflows to the community. Recently, we developed CobraMod - a pathway-centric curation tool for constraint-based metabolic models.
